Table of Links
2. Methods
2.1. Modeling Panspermia and Terraformation
2.2. Identifying the Presence of Terraformed Planets and 2.3. Software and Availability
3. Results
3.1. Panspermia can increase the correlation between planets’ compositions and positions
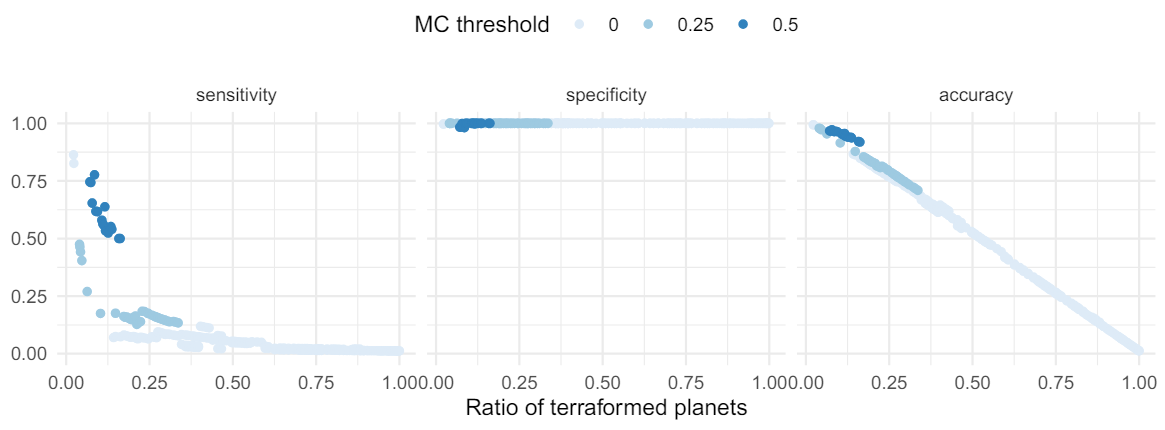
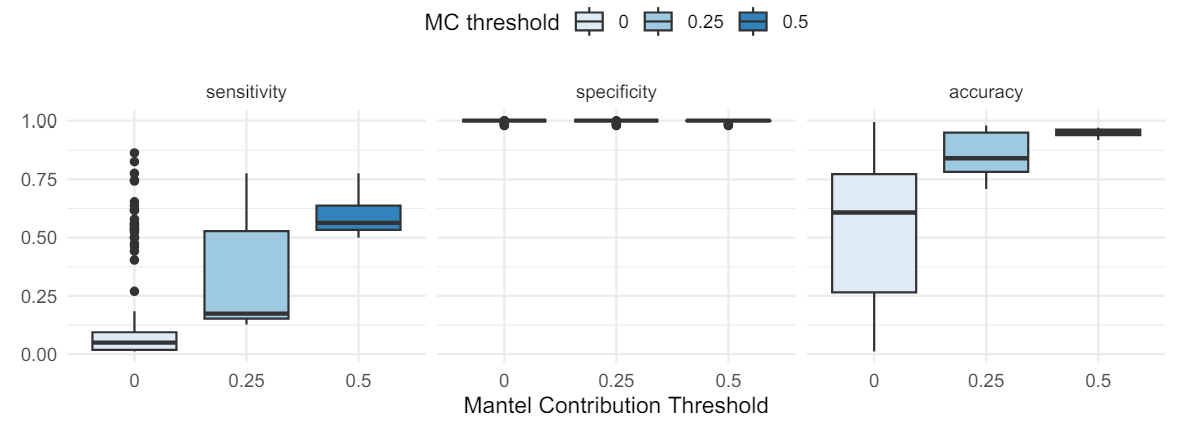
3.2. Likely terraformed planets can be identified from clustering
5. Acknowledgements and References
APPENDIX
4. SUMMARY AND DISCUSSION
We showed how a positive Mantel correlation coefficient with a low p-value can be used to identify when a region of space contains life, under the postulates that life can spread between planetary systems and terraform planets. Going beyond (or below) a population-scale biosignature, we demonstrated how to select, from a population of planets, clusters which have high-likelihood to contain terraformed planets. More importantly, these selected clusters have a low-likelihood to contain non-terraformed planets. This is increasingly important as the field of astrobiology reconciles with unavoidable false positives for more traditional biosignatures.
We assumed that the background non-terraformed distribution of planetary compositions is uncorrelated and drawn from a flat random distribution of compositions (a reasonable prior given the high sensitivity of planet formation to initial conditions of the planetary disk; (Tasker et al. 2020; Pacetti et al. 2022)). In the event that underlying abiotic planets are correlated in position and composition space (perhaps from some undiscovered astrophysical phenomena), then the value of the Mantel correlation coefficient considered to be a statistically significant biosignature would have to increase. It’s possible that if the underlying coefficient of non-terraformed planets were too high, the spread of life would actually increase the Mantel p-value, even early in the simulation. If planetary compositions are actually clustered around a few characteristic planet types, we could represent the initial non-terraformed planets as being drawn from a distribution punctuated with narrow Gaussians in composition space, and the results might be different yet. Our results underscore the importance of better understanding the baseline abiotic diversity of planetary populations, including better constraining which features of planet formation best predict planetary characteristics over time. Our model demonstrates that such information could be used to aid life detection in the absence of making progress on a theory of living systems.
Our approach here also provides an alternative to specific chemical biosignatures at the level of planets, showing the promise of detecting anomalous features at the population-scale. Critically, these anomalous features can be explained by a model with simple hypotheses about what life does, rather than what life is, and is agnostic to our ignorance of living systems as they might exist outside of Earth.
This is only an introduction to the kinds of questions which could be investigated with this technique. The basic idea of taking key features of life, and understanding how they might manifest on the scale of planets leads us to imagine properties of life that may manifest across scales, like “planetary phylogenies”—that is, heredities and lineages of planets—and how they could shed light
on better understanding living systems generally. Such hypothetical characteristics of planets (and life) could be investigated with our of model.
Perhaps one of the biggest limitations in our model is assuming we can reliably map observable characteristics of a planet to something as comprehensible and malleable as a vector of real numbers. For example, how the chemistry of the atmosphere, biosphere, geosphere, etc. could be reflected via the atmosphere of a planet, and what the mappings are from observed atmospheric properties (the planet’s “phenotype”) to the underlying chemistries that put constraints on the compatibility of planets (the planet’s “genotype”), and the kinds of life which can emerge on or coexist with a planet. This would further strengthen the realism of our agnostic approach with minimal assumptions, and in future work we would like to explore these ideas more.
While we only walk through a simulation under a single set of parameters here, our goal was to identify a best possible case scenario for applying a technique where we assume life should be able to drive correlations in planetary position-composition space based on hallmarks of life like proliferation and environmental bi-directional feedback. Our model results show promise that life could be detected at the scale of a population of planets, using information from only ≈ 1000 (perhaps fewer) planetary atmospheres, even in the absence of any information about what kinds of planetary environments are most suitable to life, or without knowing anything about the origins of life, or the peculiarities of life’s metabolic outputs. We showed how our distributed biosignature can be further refined to detect specific clusters of terraformed planets in this space, even when they only comprise a few percent of all planets. Again, this approach does not require an independent way to “detect” life, like a smoking gun biosignature atmospheric gas. Instead this approach depends on two key assumptions about what life can do, and derives observable consequences directly from them, providing a statistical approach which can be refined by astronomical surveys, and therein lies its biggest promise.
Authors:
(1) Harrison B. Smith, Earth-Life Science Institute, Tokyo Institute of Technology, Ookayama, Meguro-ku, Tokyo, Japan, and Blue Marble Space Institute of Science, Seattle, Washington, USA ([email protected]);
(2) Lana Sinapayen, Sony Computer Science Laboratories, Kyoto, Japan and National Institute for Basic Biology, Okazaki, Japan ([email protected]).
This paper is
